DisEMBL 1.5 - Predictors of intrinsic protein disorder
OVERVIEW
DIS.EMBL.DE RANKINGS
Date Range
Date Range
Date Range
LINKS TO BUSINESS
Bioinformatics has evolved as a great tool for molecular biologists. There are various tools available for helping in reducing the time required to analyze biological materials be it DNA, RNA, Proteins etc. I wish to list here few of the commonly used tools. Please send me suggestions to improve the content. If you like or dislike something, let me know, your inputs matters. Monday, October 23, 2017.
Photos of Barton Group Events. Archive of Group News 1999-2013. Archive of Group News May 1994-1999. All Barton Group Software and Databases. Dundee Resource for Sequence Analysis and Structure Prediction. Multiple Sequence Alignment and Analysis.
Genetic variants in cryptic choleostasis. Genetic variants in cryptic choleostasis. Genetic variants in cryptic choleostasis. Functional genetic variants in healthy genomes. Functional genetic variants in healthy genomes. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms.
Is supported by BMBS COST Action BM1405. COST is supported by the EU Framework Programme Horizon 2020.
Sequence sites, features and motifs. ExPASy is the SIB Bioinformatics Resource Portal. In different areas of life sciences including proteomics, genomics, phylogeny, systems biology, population genetics, transcriptomics etc. This service provides topology and parameters for small molecules, for use with CHARMM and GROMAC.
Intrinsic Protein Disorder, Domain and Globularity Prediction.
Genetic variants in cryptic choleostasis. Genetic variants in cryptic choleostasis. Genetic variants in cryptic choleostasis. Functional genetic variants in healthy genomes. Functional genetic variants in healthy genomes. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms. Cancer severity from interaction mechanisms.
Welcome by Dr Rune Linding. Introduction to the lab by Professor Rune Linding. The Linding lab is a big data network biology research group. We explore biological systems by developing and deploying. Algorithms aimed to forecast cell behaviour with an accuracy similar to that of weather or aircraft models. Our focus is on studying cellular signal processing and decision making. We work with other international integrative network, cancer and tumor biology laboratories in large-sc.
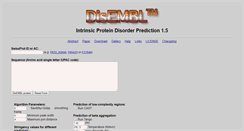
WHAT DOES DIS.EMBL.DE LOOK LIKE?

DIS.EMBL.DE HOST
BOOKMARK ICON

SERVER OPERATING SYSTEM AND ENCODING
I caught that dis.embl.de is weilding the nginx server.TITLE
DisEMBL 1.5 - Predictors of intrinsic protein disorderDESCRIPTION
Intrinsic Protein Disorder Prediction 1.5. SwissProt ID or AC. Sequence Amino acid single letter IUPAC code. Stringency values for different predictors. Use Java applet for displaying results. Prediction of low-complexity regions. Prediction of beta aggregation. R Linding, L.J. Jensen, F. Diella, P. Bork, T.J. Gibson and R.B. Russell. Protein disorder prediction implications for structural proteomics. Structure Vol 11, Issue 11, 4 November 2003. Please see the DisEMBL preview paper.CONTENT
This web page states the following, "Sequence Amino acid single letter IUPAC code." Our analyzers saw that the web site said " Stringency values for different predictors." The Website also said " Use Java applet for displaying results. Protein disorder prediction implications for structural proteomics. Structure Vol 11, Issue 11, 4 November 2003. Please see the DisEMBL preview paper." The website's header had protein disorder domain boundaries predictor prediction loops unstructured unfolded intrinsic intrinsically genomics proteomics globularity non-globular X-ray nmr polypeptides biopolymers IUP IUPs IDP IDPs ELM ELMs crystallisation construct purification expression amyloid partially folded as the most important optimized keyword.SUBSEQUENT BUSINESSES
DIS Bölüm Bilgi Sistemi, Bilgi İşlem Daire Başkanlığı.
Анализ академической среды как места учебы и работы. На протяжении последних лет наблюдается значительный рост численности студентов, которые едут учиться за рубеж, и ко.